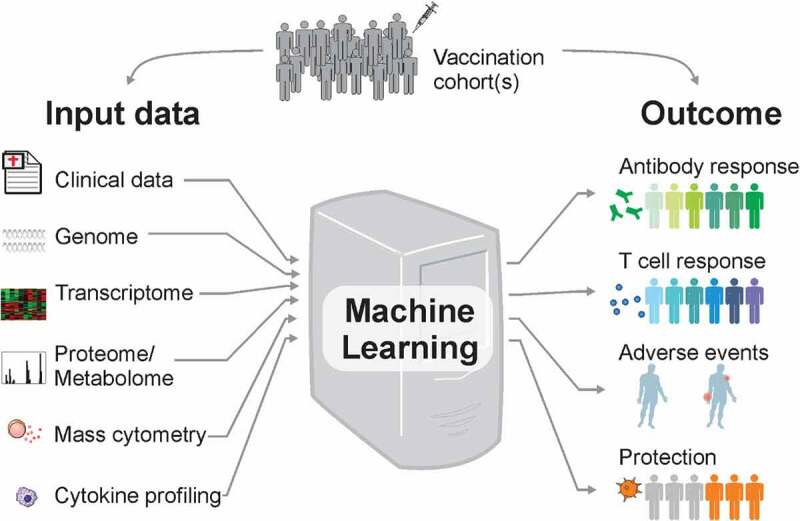
Methods for predicting vaccine immunogenicity and reactogenicity.
Subjects receiving the same vaccine often show different levels of immune responses and some may even present adverse side effects to the vaccine. Systems vaccinology can combine omics data and machine learning techniques to obtain highly predictive signatures of vaccine immunogenicity and reactogenicity. Currently, several machine learning methods are already available to researchers with no background in bioinformatics. Here we described the four main steps to discover markers of vaccine immunogenicity and reactogenicity: (1) Preparing the data; (2) Selecting the vaccinees and relevant genes; (3) Choosing the algorithm; (4) Blind testing your model. With the increasing number of Systems Vaccinology datasets being generated, we expect that the accuracy and robustness of signatures of vaccine reactogenicity and immunogenicity will significantly improve.
Authors
Patrícia Gonzalez-Dias; Eva K Lee; Sara Sorgi; Diógenes S de Lima; Alysson H Urbanski; Eduardo Lv Silveira; Helder I Nakaya
External link
Publication Year
Publication Journal
Associeted Project
Systems Vaccinology
Lista de serviços
-
Is the gut microbiome key to modulating vaccine efficacy?Is the gut microbiome key to modulating vaccine efficacy?
-
Toxicogenomic and bioinformatics platforms to identify key molecular mechanisms of a curcumin-analogue DM-1 toxicity in melanoma cells.Toxicogenomic and bioinformatics platforms to identify key molecular mechanisms of a curcumin-analogue DM-1 toxicity in melanoma cells.