Gene regulatory networks analysis for the discovery of prognostic genes in gliomas.
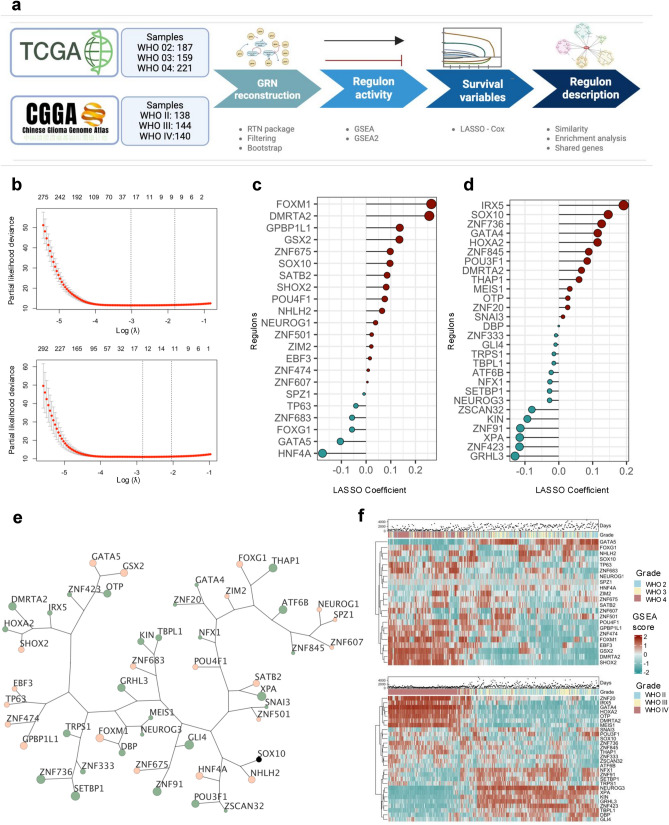
Gliomas are the most common and aggressive primary tumors of the central nervous system. Dysregulated transcription factors (TFs) and genes have been implicated in glioma progression, yet these tumors' overall structure of gene regulatory networks (GRNs) remains undefined. We analyzed transcriptional data from 989 primary gliomas in The Cancer Genome Atlas (TCGA) and the Chinese Glioma Genome Atlas (CGGA) to address this. GRNs were reconstructed using the RTN package which identifies regulons-sets of genes regulated by a common TF based on co-expression and mutual information. Regulon activity was evaluated through Gene Set Enrichment Analysis. Elastic net regularization and Cox regression identified 31 and 32 prognostic genes in the TCGA and CGGA datasets, respectively, with 11 genes overlapping, many of which are associated with neural development and synaptic processes. GAS2L3, HOXD13, and OTP demonstrated the strongest correlations with survival outcomes among these. Single-cell RNA-seq analysis of 201,986 cells revealed distinct expression patterns for these genes in glioma subpopulations, particularly oligoprogenitor cells. This study uncovers key GRNs and prognostic genes in gliomas, offering new insights into tumor biology and potential therapeutic targets.
Authors
Barcelos PM, Filgueiras IS, Nóbile AL, Usuda JN, Adri AS, et al.
External link
Publication Year
Publication Journal
Associeted Project
Network & Precision Medicine
Lista de serviços
-
Is the gut microbiome key to modulating vaccine efficacy?Is the gut microbiome key to modulating vaccine efficacy?
-
Toxicogenomic and bioinformatics platforms to identify key molecular mechanisms of a curcumin-analogue DM-1 toxicity in melanoma cells.Toxicogenomic and bioinformatics platforms to identify key molecular mechanisms of a curcumin-analogue DM-1 toxicity in melanoma cells.