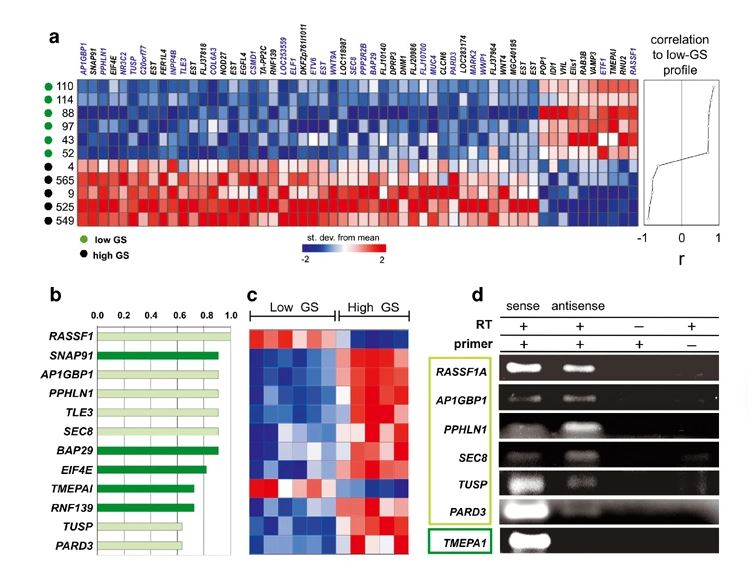
Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer.
A large fraction of transcripts are expressed antisense to introns of known genes in the human genome. Here we show the construction and use of a cDNA microarray platform enriched in intronic transcripts to assess their biological relevance in pathological conditions. To validate the approach, prostate cancer was used as a model, and 27 patient tumor samples with Gleason scores ranging from 5 to 10 were analyzed. We find that a considerably higher fraction (6.6%, [23/346]) of intronic transcripts are significantly correlated (P< or =0.001) to the degree of prostate tumor differentiation (Gleason score) when compared to transcripts from unannotated genomic regions (1%, [6/539]) or from exons of known genes (2%, [27/1369]). Among the top twelve transcripts most correlated to tumor differentiation, six are antisense intronic messages as shown by orientation-specific RT-PCR or Northern blot analysis with strand-specific riboprobe. Orientation-specific real-time RT-PCR with six tumor samples, confirmed the correlation (P=0.024) between the low/high degrees of tumor differentiation and antisense intronic RASSF1 transcript levels. The need to use intron arrays to reveal the transcriptome profile of antisense intronic RNA in cancer has clearly emerged.
Authors
Eduardo M Reis; Helder I Nakaya; Rodrigo Louro; Flavio C Canavez; Aurea V F Flatschart; Giulliana T Almeida; Camila M Egidio; Apuã C Paquola; Abimael A Machado; Fernanda Festa; Denise Yamamoto; Renato Alvarenga; Camille C da Silva; Glauber C Brito; Sérgio D Simon; Carlos A Moreira-Filho; Katia R Leite; Luiz H Camara-Lopes; Franz S Campos; Etel Gimba; Giselle M Vignal; Hamza El-Dorry; Mari C Sogayar; Marcello A Barcinski; Aline M da Silva; Sergio Verjovski-Almeida
External link
Publication Year
Publication Journal
Associeted Project
Long Noncoding RNAs
Lista de serviços
-
Gene regulatory and signaling networks exhibit distinct topological distributions of motifs.Gene regulatory and signaling networks exhibit distinct topological distributions of motifs.
-
Gene signatures of autopsy lungs from obese patients with COVID-19.Gene signatures of autopsy lungs from obese patients with COVID-19.
-
Network Medicine: Methods and ApplicationsNetwork Medicine: Methods and Applications
-
ACE2 Expression Is Increased in the Lungs of Patients With Comorbidities Associated With Severe COVID-19.ACE2 Expression Is Increased in the Lungs of Patients With Comorbidities Associated With Severe COVID-19.
-
Drug repositioning for psychiatric and neurological disorders through a network medicine approach.Drug repositioning for psychiatric and neurological disorders through a network medicine approach.
-
Linking proteomic alterations in schizophrenia hippocampus to NMDAr hypofunction in human neurons and oligodendrocytes.Linking proteomic alterations in schizophrenia hippocampus to NMDAr hypofunction in human neurons and oligodendrocytes.
-
In-depth analysis of laboratory parameters reveals the interplay between sex, age, and systemic inflammation in individuals with COVID-19.In-depth analysis of laboratory parameters reveals the interplay between sex, age, and systemic inflammation in individuals with COVID-19.
-
The evolution of knowledge on genes associated with human diseasesThe evolution of knowledge on genes associated with human diseases
-
Network vaccinology.Network vaccinology.
-
Pyruvate kinase M2 mediates IL-17 signaling in keratinocytes driving psoriatic skin inflammationPyruvate kinase M2 mediates IL-17 signaling in keratinocytes driving psoriatic skin inflammation
-
Transcriptome analysis of six tissues obtained post-mortem from sepsis patientsTranscriptome analysis of six tissues obtained post-mortem from sepsis patients
-
Gene Signatures of Symptomatic and Asymptomatic Clinical-Immunological Profiles of Human Infection by Leishmania (L.) chagasi in Amazonian BrazilGene Signatures of Symptomatic and Asymptomatic Clinical-Immunological Profiles of Human Infection by Leishmania (L.) chagasi in Amazonian Brazil
-
In vitro morphological profiling of T cells predicts clinical response to natalizumab therapy in patients with multiple sclerosis.In vitro morphological profiling of T cells predicts clinical response to natalizumab therapy in patients with multiple sclerosis.
-
Integrative immunology identified interferome signatures in uveitis and systemic disease-associated uveitis.Integrative immunology identified interferome signatures in uveitis and systemic disease-associated uveitis.
-
Gene regulatory networks analysis for the discovery of prognostic genes in gliomas.Gene regulatory networks analysis for the discovery of prognostic genes in gliomas.
-
Revealing shared molecular drivers of brain metastases from distinct primary tumors.Revealing shared molecular drivers of brain metastases from distinct primary tumors.