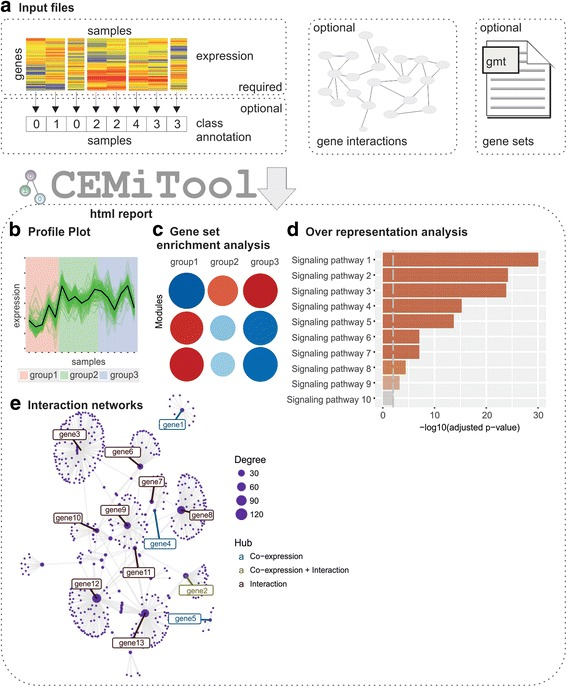
CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.
The analysis of modular gene co-expression networks is a well-established method commonly used for discovering the systems-level functionality of genes. In addition, these studies provide a basis for the discovery of clinically relevant molecular pathways underlying different diseases and conditions. In this paper, we present a fast and easy-to-use Bioconductor package named CEMiTool that unifies the discovery and the analysis of co-expression modules. Using the same real datasets, we demonstrate that CEMiTool outperforms existing tools, and provides unique results in a user-friendly html report with high quality graphs. Among its features, our tool evaluates whether modules contain genes that are over-represented by specific pathways or that are altered in a specific sample group, as well as it integrates transcriptomic data with interactome information, identifying the potential hubs on each network. We successfully applied CEMiTool to over 1000 transcriptome datasets, and to a new RNA-seq dataset of patients infected with Leishmania, revealing novel insights of the disease's physiopathology. The CEMiTool R package provides users with an easy-to-use method to automatically implement gene co-expression network analyses, obtain key information about the discovered gene modules using additional downstream analyses and retrieve publication-ready results via a high-quality interactive report.
Authors
Pedro S T Russo; Gustavo R Ferreira; Lucas E Cardozo; Matheus C Bürger; Raul Arias-Carrasco; Sandra R Maruyama; Thiago D C Hirata; Diógenes S Lima; Fernando M Passos; Kiyoshi F Fukutani; Melissa Lever; João S Silva; Vinicius Maracaja-Coutinho; Helder I Nakaya
External link
Publication Year
Publication Journal
Associeted Project
User-friendly computational Tools
Lista de serviços
-
Gene regulatory and signaling networks exhibit distinct topological distributions of motifs.Gene regulatory and signaling networks exhibit distinct topological distributions of motifs.
-
Gene signatures of autopsy lungs from obese patients with COVID-19.Gene signatures of autopsy lungs from obese patients with COVID-19.
-
Network Medicine: Methods and ApplicationsNetwork Medicine: Methods and Applications
-
ACE2 Expression Is Increased in the Lungs of Patients With Comorbidities Associated With Severe COVID-19.ACE2 Expression Is Increased in the Lungs of Patients With Comorbidities Associated With Severe COVID-19.
-
Drug repositioning for psychiatric and neurological disorders through a network medicine approach.Drug repositioning for psychiatric and neurological disorders through a network medicine approach.
-
Linking proteomic alterations in schizophrenia hippocampus to NMDAr hypofunction in human neurons and oligodendrocytes.Linking proteomic alterations in schizophrenia hippocampus to NMDAr hypofunction in human neurons and oligodendrocytes.
-
In-depth analysis of laboratory parameters reveals the interplay between sex, age, and systemic inflammation in individuals with COVID-19.In-depth analysis of laboratory parameters reveals the interplay between sex, age, and systemic inflammation in individuals with COVID-19.
-
The evolution of knowledge on genes associated with human diseasesThe evolution of knowledge on genes associated with human diseases
-
Network vaccinology.Network vaccinology.
-
Pyruvate kinase M2 mediates IL-17 signaling in keratinocytes driving psoriatic skin inflammationPyruvate kinase M2 mediates IL-17 signaling in keratinocytes driving psoriatic skin inflammation
-
Transcriptome analysis of six tissues obtained post-mortem from sepsis patientsTranscriptome analysis of six tissues obtained post-mortem from sepsis patients
-
Gene Signatures of Symptomatic and Asymptomatic Clinical-Immunological Profiles of Human Infection by Leishmania (L.) chagasi in Amazonian BrazilGene Signatures of Symptomatic and Asymptomatic Clinical-Immunological Profiles of Human Infection by Leishmania (L.) chagasi in Amazonian Brazil
-
In vitro morphological profiling of T cells predicts clinical response to natalizumab therapy in patients with multiple sclerosis.In vitro morphological profiling of T cells predicts clinical response to natalizumab therapy in patients with multiple sclerosis.
-
Integrative immunology identified interferome signatures in uveitis and systemic disease-associated uveitis.Integrative immunology identified interferome signatures in uveitis and systemic disease-associated uveitis.
-
Gene regulatory networks analysis for the discovery of prognostic genes in gliomas.Gene regulatory networks analysis for the discovery of prognostic genes in gliomas.
-
Revealing shared molecular drivers of brain metastases from distinct primary tumors.Revealing shared molecular drivers of brain metastases from distinct primary tumors.