SARS-CoV-2 Selectively Induces the Expression of Unproductive Splicing Isoforms of Interferon, Class I MHC, and Splicing Machinery Genes.
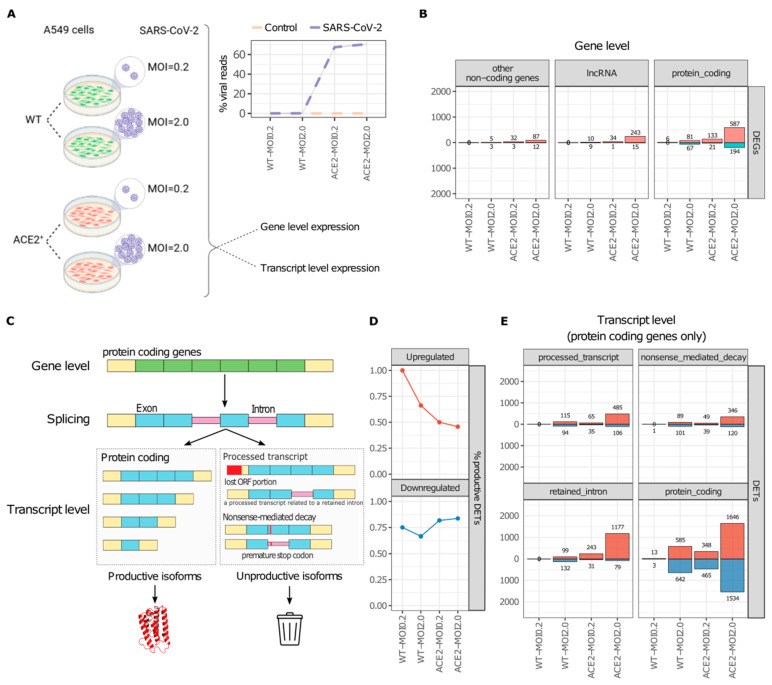
RNA processing is a highly conserved mechanism that serves as a pivotal regulator of gene expression. Alternative processing generates transcripts that can still be translated but lead to potentially nonfunctional proteins. A plethora of respiratory viruses, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), strategically manipulate the host's RNA processing machinery to circumvent antiviral responses. We integrated publicly available omics datasets to systematically analyze isoform-level expression and delineate the nascent peptide landscape of SARS-CoV-2-infected human cells. Our findings explore a suggested but uncharacterized mechanism, whereby SARS-CoV-2 infection induces the predominant expression of unproductive splicing isoforms in key IFN signaling, interferon-stimulated (ISGs), class I MHC, and splicing machinery genes, including IRF7, HLA-B, and HNRNPH1. In stark contrast, cytokine and chemokine genes, such as IL6 and TNF, predominantly express productive (protein-coding) splicing isoforms in response to SARS-CoV-2 infection. We postulate that SARS-CoV-2 employs an unreported tactic of exploiting the host splicing machinery to bolster viral replication and subvert the immune response by selectively upregulating unproductive splicing isoforms from antigen presentation and antiviral response genes. Our study sheds new light on the molecular interplay between SARS-CoV-2 and the host immune system, offering a foundation for the development of novel therapeutic strategies to combat COVID-19.
Authors
Dias TL, Mamede I, de Toledo NE, Queiroz LR
External link
Publication Year
Publication Journal
Associeted Project
Long Noncoding RNAs
Lista de serviços
-
Gene regulatory and signaling networks exhibit distinct topological distributions of motifs.Gene regulatory and signaling networks exhibit distinct topological distributions of motifs.
-
Gene signatures of autopsy lungs from obese patients with COVID-19.Gene signatures of autopsy lungs from obese patients with COVID-19.
-
Network Medicine: Methods and ApplicationsNetwork Medicine: Methods and Applications
-
ACE2 Expression Is Increased in the Lungs of Patients With Comorbidities Associated With Severe COVID-19.ACE2 Expression Is Increased in the Lungs of Patients With Comorbidities Associated With Severe COVID-19.
-
Drug repositioning for psychiatric and neurological disorders through a network medicine approach.Drug repositioning for psychiatric and neurological disorders through a network medicine approach.
-
Linking proteomic alterations in schizophrenia hippocampus to NMDAr hypofunction in human neurons and oligodendrocytes.Linking proteomic alterations in schizophrenia hippocampus to NMDAr hypofunction in human neurons and oligodendrocytes.
-
In-depth analysis of laboratory parameters reveals the interplay between sex, age, and systemic inflammation in individuals with COVID-19.In-depth analysis of laboratory parameters reveals the interplay between sex, age, and systemic inflammation in individuals with COVID-19.
-
The evolution of knowledge on genes associated with human diseasesThe evolution of knowledge on genes associated with human diseases
-
Network vaccinology.Network vaccinology.
-
Pyruvate kinase M2 mediates IL-17 signaling in keratinocytes driving psoriatic skin inflammationPyruvate kinase M2 mediates IL-17 signaling in keratinocytes driving psoriatic skin inflammation
-
Transcriptome analysis of six tissues obtained post-mortem from sepsis patientsTranscriptome analysis of six tissues obtained post-mortem from sepsis patients
-
Gene Signatures of Symptomatic and Asymptomatic Clinical-Immunological Profiles of Human Infection by Leishmania (L.) chagasi in Amazonian BrazilGene Signatures of Symptomatic and Asymptomatic Clinical-Immunological Profiles of Human Infection by Leishmania (L.) chagasi in Amazonian Brazil
-
In vitro morphological profiling of T cells predicts clinical response to natalizumab therapy in patients with multiple sclerosis.In vitro morphological profiling of T cells predicts clinical response to natalizumab therapy in patients with multiple sclerosis.
-
Integrative immunology identified interferome signatures in uveitis and systemic disease-associated uveitis.Integrative immunology identified interferome signatures in uveitis and systemic disease-associated uveitis.
-
Gene regulatory networks analysis for the discovery of prognostic genes in gliomas.Gene regulatory networks analysis for the discovery of prognostic genes in gliomas.
-
Revealing shared molecular drivers of brain metastases from distinct primary tumors.Revealing shared molecular drivers of brain metastases from distinct primary tumors.