Phenotype, function, and gene expression profiles of programmed death-1(hi) CD8 T cells in healthy human adults.
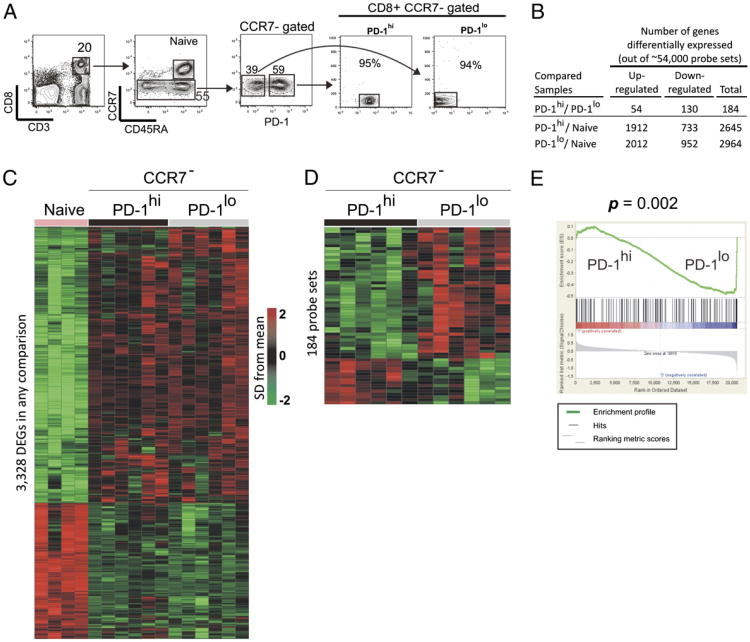
T cell dysfunction is an important feature of many chronic viral infections. In particular, it was shown that programmed death-1 (PD-1) regulates T cell dysfunction during chronic lymphocytic choriomeningitis virus infection in mice, and PD-1(hi) cells exhibit an intense exhausted gene signature. These findings were extended to human chronic infections such as HIV, hepatitis C virus, and hepatitis B virus. However, it is not known if PD-1(hi) cells of healthy humans have the traits of exhausted cells. In this study, we provide a comprehensive description of phenotype, function, and gene expression profiles of PD-1(hi) versus PD-1(lo) CD8 T cells in the peripheral blood of healthy human adults as follows: 1) the percentage of naive and memory CD8 T cells varied widely in the peripheral blood cells of healthy humans, and PD-1 was expressed by the memory CD8 T cells; 2) PD-1(hi) CD8 T cells in healthy humans did not significantly correlate with the PD-1(hi) exhausted gene signature of HIV-specific human CD8 T cells or chronic lymphocytic choriomeningitis virus-specific CD8 T cells from mice; 3) PD-1 expression did not directly affect the ability of CD8 T cells to secrete cytokines in healthy adults; 4) PD-1 was expressed by the effector memory compared with terminally differentiated effector CD8 T cells; and 5) finally, an interesting inverse relationship between CD45RA and PD-1 expression was observed. In conclusion, our study shows that most PD-1(hi) CD8 T cells in healthy adult humans are effector memory cells rather than exhausted cells.
Authors
Jaikumar Duraiswamy; Chris C Ibegbu; David Masopust; Joseph D Miller; Koichi Araki; Gregory H Doho; Pramila Tata; Satish Gupta; Michael J Zilliox; Helder I Nakaya; Bali Pulendran; W Nicholas Haining; Gordon J Freeman; Rafi Ahmed
External link
Publication Year
Publication Journal
Associeted Project
Microbiology or Immunology
Lista de serviços
-
StructRNAfinder: an automated pipeline and web server for RNA families prediction.StructRNAfinder: an automated pipeline and web server for RNA families prediction.
-
CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.
-
webCEMiTool: Co-expression Modular Analysis Made Easy.webCEMiTool: Co-expression Modular Analysis Made Easy.
-
Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.
-
Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.
-
OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.
-
Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.
-
Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.
-
Automatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone imagesAutomatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone images
-
Tucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approachTucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approach
-
Ten quick tips for harnessing the power of ChatGPT in computational biologyTen quick tips for harnessing the power of ChatGPT in computational biology