Systems vaccinology: its promise and challenge for HIV vaccine development.
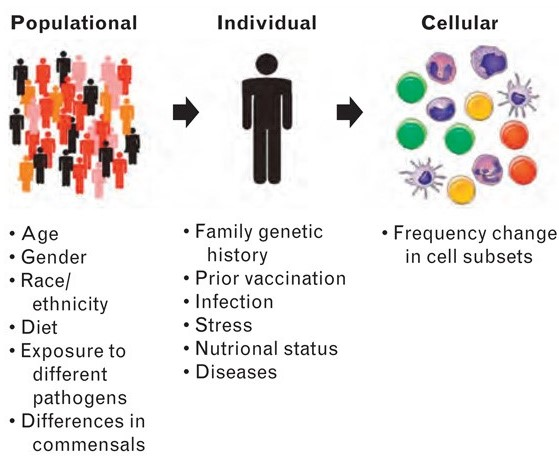
The use of systems biology approaches to understand and predict vaccine-induced immunity promises to revolutionize vaccinology. For centuries vaccines were developed empirically, with very little understanding of the mechanisms by which they mediate protective immunity. The so-called systems vaccinology approach employs high-throughput technologies (e.g. microarrays, RNA-seq and mass spectrometry-based proteomics and metabolomics) and computational modeling to describe the complex interactions between all the parts of immune system, with a view to elucidating new biological rules capable of predicting the behavior of the system. Systems biology successfully applied to yellow-fever and influenza vaccines has led to the discovery of signatures that predict vaccine immunogenicity, and promises to advance basic immunology research by providing novel mechanistic insights about immune regulation. However a major challenge of systems vaccinology concerns the analyses and interpretation of the large and noisy data sets generated by high-throughput techniques. Overcoming these issues, we envision that systems vaccinology will have a potential impact on vaccine development, including HIV vaccines. High-throughput technologies allow the investigation of vaccine-induced immune responses at system and molecular levels. These are currently being used to unravel new molecular insights about the immune system, and are on the verge of being integrated into clinical trials to enable rational vaccine design and development.
Publication Year
Publication Journal
Associeted Project
Systems Vaccinology
Lista de serviços
-
StructRNAfinder: an automated pipeline and web server for RNA families prediction.StructRNAfinder: an automated pipeline and web server for RNA families prediction.
-
CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.
-
webCEMiTool: Co-expression Modular Analysis Made Easy.webCEMiTool: Co-expression Modular Analysis Made Easy.
-
Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.
-
Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.
-
OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.
-
Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.
-
Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.
-
Automatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone imagesAutomatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone images
-
Tucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approachTucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approach
-
Ten quick tips for harnessing the power of ChatGPT in computational biologyTen quick tips for harnessing the power of ChatGPT in computational biology