Gene profiling of Chikungunya virus arthritis in a mouse model reveals significant overlap with rheumatoid arthritis.
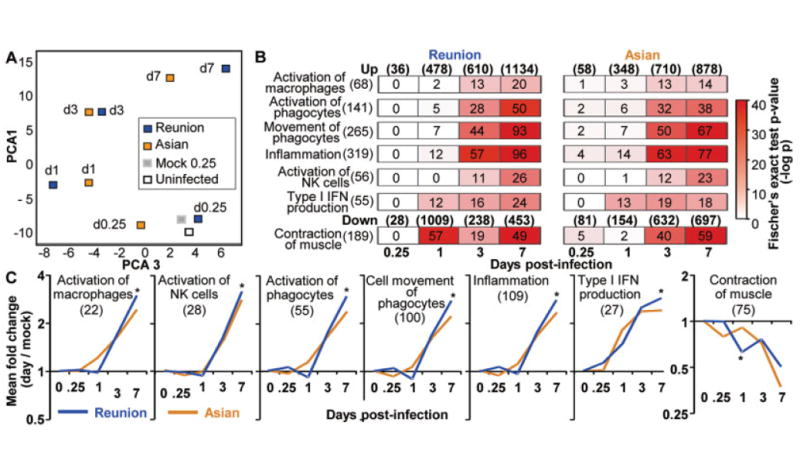
Chikungunya virus (CHIKV) is a mosquito-borne alphavirus that causes a chronic debilitating polyarthralgia/polyarthritis, for which current treatments are often inadequate. To assess whether new drugs being developed for rheumatoid arthritis (RA) might find utility in the treatment of alphaviral arthritides, we sought to determine whether the inflammatory gene expression signature of CHIKV arthritis shows any similarities with RA or collagen-induced arthritis (CIA), a mouse model of RA. Using a recently developed animal model of CHIKV arthritis in adult wild-type mice, we generated a consensus CHIKV arthritis gene expression signature, which was used to interrogate publicly available microarray studies of RA and CIA. Pathway analyses were then performed using the overlapping gene signatures. Gene set enrichment analysis showed that there was a highly significant overlap in the differentially expressed genes in the CHIKV arthritis model and in RA. This concordance also increased with the severity of RA, as measured by the inflammation score. A highly significant overlap was also seen between CHIKV arthritis and CIA. Pathway analysis revealed that the overlap between these arthritides was spread over a range of different inflammatory processes. Involvement of T cells and interferon-γ (IFNγ) in CHIKV arthritis was confirmed in studies of MHCII-deficient mice and IFNγ-deficient mice, respectively. These results suggest that RA, a chronic autoimmune arthritis, and CHIKV disease, usually a self-limiting viral arthropathy, share multiple inflammatory processes. New drugs and biologic therapies being developed for RA may thus find application in the treatment of alphaviral arthritides.
Authors
Helder I Nakaya; Joy Gardner; Yee-Suan Poo; Lee Major; Bali Pulendran; Andreas Suhrbier
External link
Publication Year
Publication Journal
Associeted Project
Systems Immunology of Human Diseases
Lista de serviços
-
StructRNAfinder: an automated pipeline and web server for RNA families prediction.StructRNAfinder: an automated pipeline and web server for RNA families prediction.
-
CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.
-
webCEMiTool: Co-expression Modular Analysis Made Easy.webCEMiTool: Co-expression Modular Analysis Made Easy.
-
Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.
-
Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.
-
OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.
-
Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.
-
Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.
-
Automatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone imagesAutomatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone images
-
Tucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approachTucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approach
-
Ten quick tips for harnessing the power of ChatGPT in computational biologyTen quick tips for harnessing the power of ChatGPT in computational biology