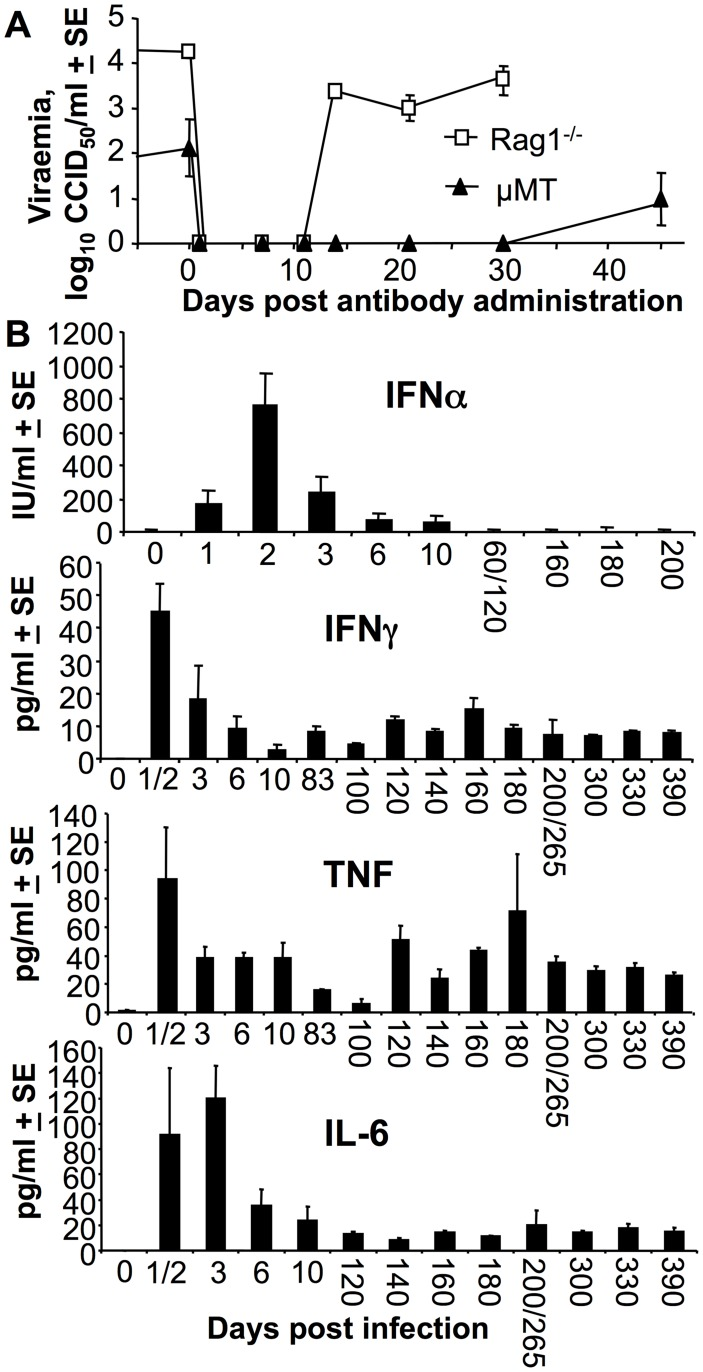
Multiple immune factors are involved in controlling acute and chronic chikungunya virus infection.
The recent epidemic of the arthritogenic alphavirus, chikungunya virus (CHIKV) has prompted a quest to understand the correlates of protection against virus and disease in order to inform development of new interventions. Herein we highlight the propensity of CHIKV infections to persist long term, both as persistent, steady-state, viraemias in multiple B cell deficient mouse strains, and as persistent RNA (including negative-strand RNA) in wild-type mice. The knockout mouse studies provided evidence for a role for T cells (but not NK cells) in viraemia suppression, and confirmed the role of T cells in arthritis promotion, with vaccine-induced T cells also shown to be arthritogenic in the absence of antibody responses. However, MHC class II-restricted T cells were not required for production of anti-viral IgG2c responses post CHIKV infection. The anti-viral cytokines, TNF and IFNγ, were persistently elevated in persistently infected B and T cell deficient mice, with adoptive transfer of anti-CHIKV antibodies unable to clear permanently the viraemia from these, or B cell deficient, mice. The NOD background increased viraemia and promoted arthritis, with B, T and NK deficient NOD mice showing high-levels of persistent viraemia and ultimately succumbing to encephalitic disease. In wild-type mice persistent CHIKV RNA and negative strand RNA (detected for up to 100 days post infection) was associated with persistence of cellular infiltrates, CHIKV antigen and stimulation of IFNα/β and T cell responses. These studies highlight that, secondary to antibodies, several factors are involved in virus control, and suggest that chronic arthritic disease is a consequence of persistent, replicating and transcriptionally active CHIKV RNA.
Authors
Yee Suan Poo; Penny A Rudd; Joy Gardner; Jane A C Wilson; Thibaut Larcher; Marie-Anne Colle; Thuy T Le; Helder I Nakaya; David Warrilow; Richard Allcock; Helle Bielefeldt-Ohmann; Wayne A Schroder; Alexander A Khromykh; José A Lopez; Andreas Suhrbier
External link
Publication Year
Publication Journal
Associeted Project
Microbiology or Immunology
Lista de serviços
-
StructRNAfinder: an automated pipeline and web server for RNA families prediction.StructRNAfinder: an automated pipeline and web server for RNA families prediction.
-
CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.
-
webCEMiTool: Co-expression Modular Analysis Made Easy.webCEMiTool: Co-expression Modular Analysis Made Easy.
-
Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.
-
Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.
-
OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.
-
Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.
-
Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.
-
Automatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone imagesAutomatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone images
-
Tucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approachTucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approach
-
Ten quick tips for harnessing the power of ChatGPT in computational biologyTen quick tips for harnessing the power of ChatGPT in computational biology