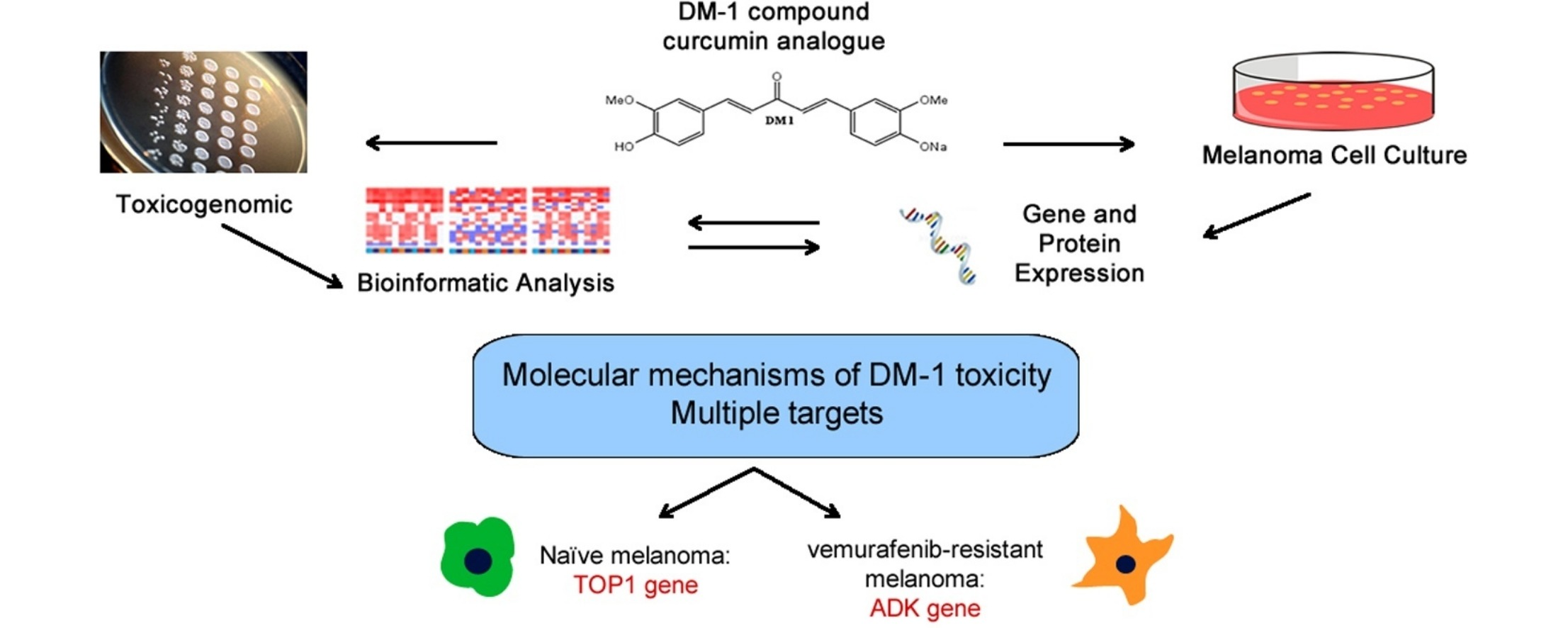
Toxicogenomic and bioinformatics platforms to identify key molecular mechanisms of a curcumin-analogue DM-1 toxicity in melanoma cells.
Melanoma is a highly invasive and metastatic cancer with high mortality rates and chemoresistance. Around 50% of melanomas are driven by activating mutations in BRAF that has led to the development of potent anti-BRAF inhibitors. However resistance to anti-BRAF therapy usually develops within a few months and consequently there is a need to identify alternative therapies that will bypass BRAF inhibitor resistance. The curcumin analogue DM-1 (sodium 4-[5-(4-hydroxy-3-methoxy-phenyl)-3-oxo-penta-1,4-dienyl]-2-methoxy-phenolate) has substantial anti-tumor activity in melanoma, but its mechanism of action remains unclear. Here we use a synthetic lethal genetic screen in Saccharomyces cerevisiae to identify 211 genes implicated in sensitivity to DM-1 toxicity. From these 211 genes, 74 had close human orthologues implicated in oxidative phosphorylation, insulin signaling and iron and RNA metabolism. Further analysis identified 7 target genes (ADK, ATP6V0B, PEMT, TOP1, ZFP36, ZFP36L1, ZFP36L2) with differential expression during melanoma progression implicated in regulation of tumor progression, cell differentiation, and epithelial-mesenchymal transition. Of these TOP1 and ADK were regulated by DM-1 in treatment-naïve and vemurafenib-resistant melanoma cells respectively. These data reveal that the anticancer effect of curcumin analogues is likely to be mediated via multiple targets and identify several genes that represent candidates for combinatorial targeting in melanoma.
Authors
Érica Aparecida de Oliveira; Diogenes Saulo de Lima; Lucas Esteves Cardozo; Garcia Ferreira de Souza; Nayane de Souza; Debora Kristina Alves-Fernandes; Fernanda Faião-Flores; José Agustín Pablo Quincoces; Silvia Berlanga de Moraes Barros; Helder I Nakaya; Gisele Monteiro; Silvya Stuchi Maria-Engler
External link
Publication Year
Publication Journal
Associeted Project
Drug repositioning
Lista de serviços
-
StructRNAfinder: an automated pipeline and web server for RNA families prediction.StructRNAfinder: an automated pipeline and web server for RNA families prediction.
-
CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.
-
webCEMiTool: Co-expression Modular Analysis Made Easy.webCEMiTool: Co-expression Modular Analysis Made Easy.
-
Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.
-
Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.
-
OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.
-
Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.
-
Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.
-
Automatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone imagesAutomatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone images
-
Tucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approachTucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approach
-
Ten quick tips for harnessing the power of ChatGPT in computational biologyTen quick tips for harnessing the power of ChatGPT in computational biology