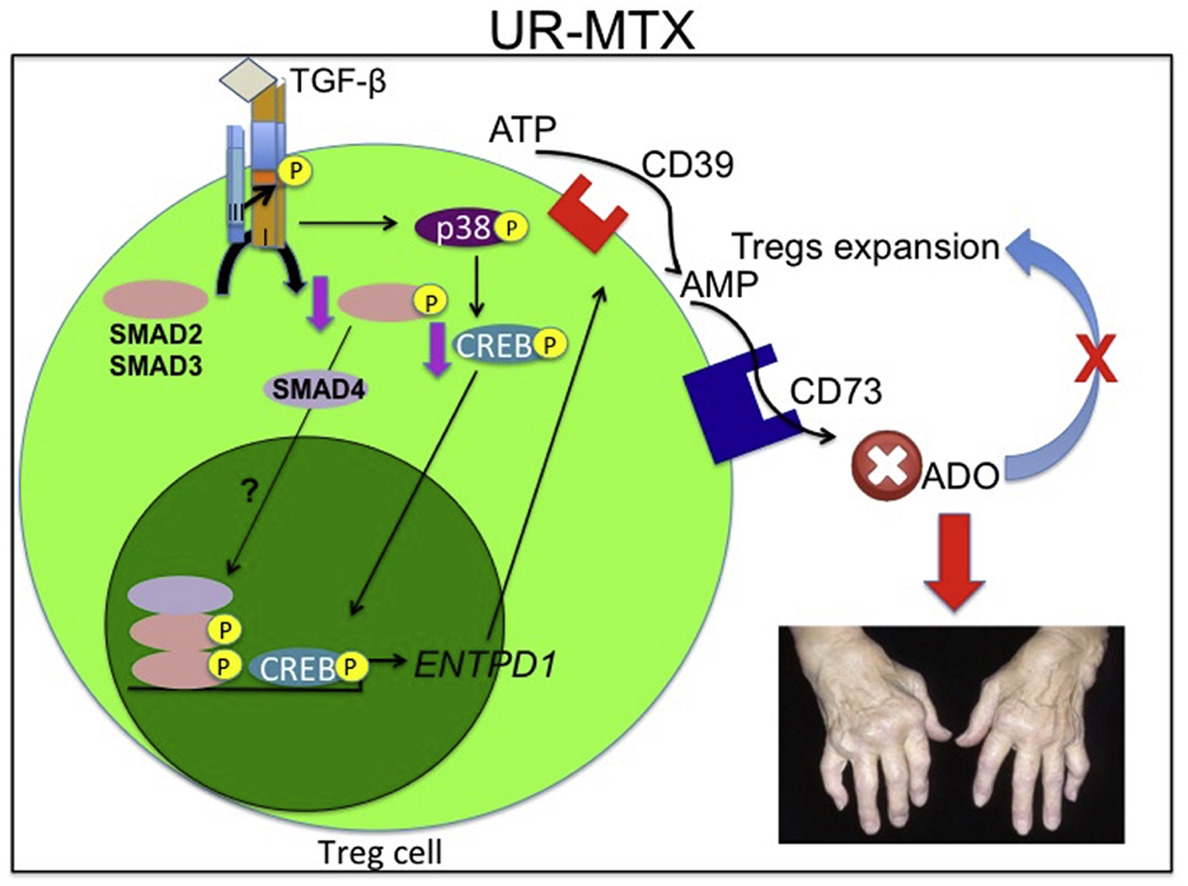
TGF-β signalling defect is linked to low CD39 expression on regulatory T cells and methotrexate resistance in rheumatoid arthritis.
Rheumatoid arthritis (RA) is an autoimmune arthropathy characterized by chronic articular inflammation. Methotrexate (MTX) remains the first-line therapy for RA and its anti-inflammatory effect is associated with the maintenance of high levels of extracellular adenosine (ADO). Nonetheless, up to 40% of RA patients are resistant to MTX treatment and this is linked to a reduction of CD39 expression, an ectoenzyme involved in the generation of extracellular ADO by ATP metabolism, on circulating regulatory T cells (Tregs). However, the mechanism mediating the reduction of CD39 expression on Tregs is unknown. Here we demonstrated that the impairment in TGF-β signalling lead to the reduction of CD39 expression on Tregs that accounts for MTX resistance. TGF-β increases CD39 expression on Tregs via the activation of TGFBRII/TGFBRI, SMAD2 and the transcription factor CREB, which is activated in a p38-dependent manner and induces CD39 expression by promoting ENTPD1 gene transcription. Importantly, unresponsive patients to MTX (UR-MTX) show reduced expression of TGFBR2 and CREB1 and decreased levels of p-SMAD2 and p-CREB in Tregs compared to MTX-responsive patients (R-MTX). Furthermore, RA patients carrying at least one mutant allele for rs1431131 (AT or AA) of the TGFBR2 gene are significantly (p = 0.0006) associated with UR-MTX. Therefore, we have uncovered a molecular mechanism for the reduced CD39 expression on Tregs, and revealed potential targets for therapeutic intervention for MTX resistance.
Authors
Raphael S Peres; Paula B Donate; Jhimmy Talbot; Nerry T Cecilio; Patricia R Lobo; Caio C Machado; Kalil W A Lima; Rene D Oliveira; Vanessa Carregaro; Helder I Nakaya; Thiago M Cunha; José Carlos Alves-Filho; Foo Y Liew; Paulo Louzada-Junior; Fernando Q Cunha
External link
Publication Year
Publication Journal
Associeted Project
Microbiology or Immunology
Lista de serviços
-
StructRNAfinder: an automated pipeline and web server for RNA families prediction.StructRNAfinder: an automated pipeline and web server for RNA families prediction.
-
CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.
-
webCEMiTool: Co-expression Modular Analysis Made Easy.webCEMiTool: Co-expression Modular Analysis Made Easy.
-
Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.
-
Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.
-
OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.
-
Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.
-
Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.
-
Automatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone imagesAutomatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone images
-
Tucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approachTucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approach
-
Ten quick tips for harnessing the power of ChatGPT in computational biologyTen quick tips for harnessing the power of ChatGPT in computational biology