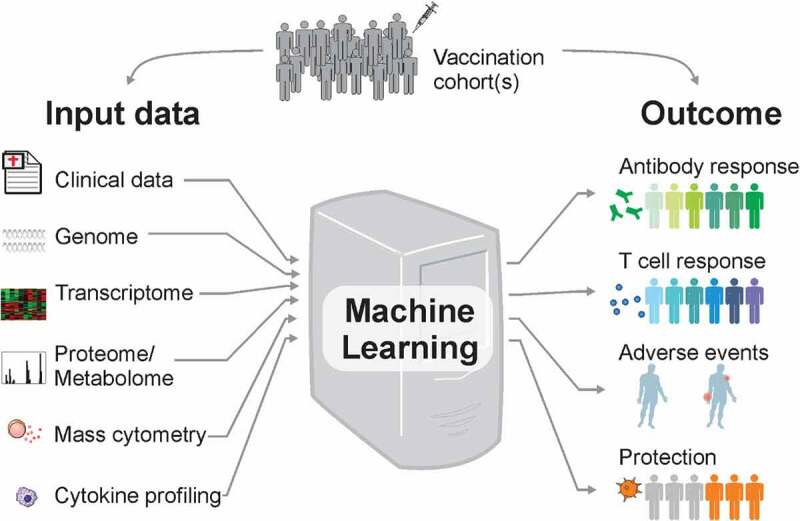
Methods for predicting vaccine immunogenicity and reactogenicity.
Subjects receiving the same vaccine often show different levels of immune responses and some may even present adverse side effects to the vaccine. Systems vaccinology can combine omics data and machine learning techniques to obtain highly predictive signatures of vaccine immunogenicity and reactogenicity. Currently, several machine learning methods are already available to researchers with no background in bioinformatics. Here we described the four main steps to discover markers of vaccine immunogenicity and reactogenicity: (1) Preparing the data; (2) Selecting the vaccinees and relevant genes; (3) Choosing the algorithm; (4) Blind testing your model. With the increasing number of Systems Vaccinology datasets being generated, we expect that the accuracy and robustness of signatures of vaccine reactogenicity and immunogenicity will significantly improve.
Authors
Patrícia Gonzalez-Dias; Eva K Lee; Sara Sorgi; Diógenes S de Lima; Alysson H Urbanski; Eduardo Lv Silveira; Helder I Nakaya
External link
Publication Year
Publication Journal
Associeted Project
Systems Vaccinology
Lista de serviços
-
StructRNAfinder: an automated pipeline and web server for RNA families prediction.StructRNAfinder: an automated pipeline and web server for RNA families prediction.
-
CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.
-
webCEMiTool: Co-expression Modular Analysis Made Easy.webCEMiTool: Co-expression Modular Analysis Made Easy.
-
Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.
-
Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.
-
OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.
-
Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.
-
Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.
-
Automatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone imagesAutomatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone images
-
Tucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approachTucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approach
-
Ten quick tips for harnessing the power of ChatGPT in computational biologyTen quick tips for harnessing the power of ChatGPT in computational biology