Integrative immunology identified interferome signatures in uveitis and systemic disease-associated uveitis.
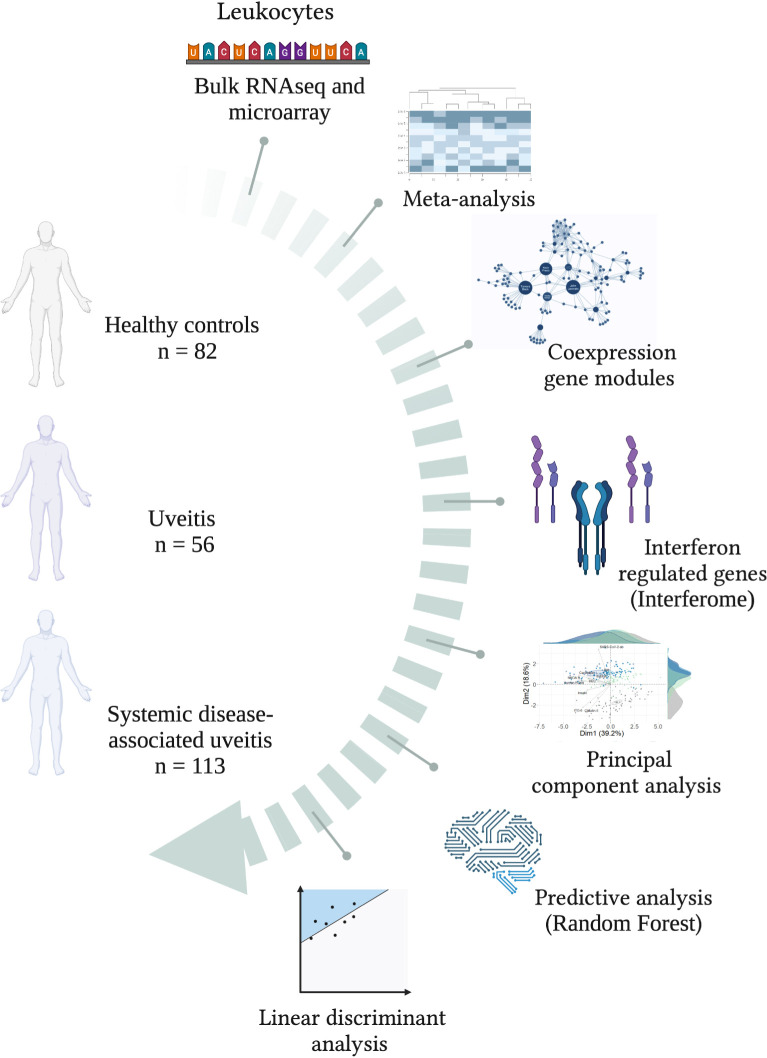
INTRODUCTION: Uveitis accounts for up to 25% of global legal blindness and involves intraocular inflammation, classifed as infectious or non-infectious. Its complex pathophysiology includes dysregulated cytokines, particularly interferons (IFNs). However, the global signature of type I, II, and III interferon-regulated genes (Interferome) remains largely uncharacterized in uveitis. METHODS: In this study, we conducted an integrative systems biology analysis of blood transcriptome data from 169 non-infectious uveitis patients (56 isolated uveitis, 113 systemic disease-associated uveitis) and 82 healthy controls. RESULTS: Modular co-expression analysis identified distinct cytokine signaling networks, emphasizing interleukin and interferon pathways. A meta-analysis revealed 110 differentially expressed genes (metaDEGs) in isolated uveitis and 91 in systemic disease-associated uveitis, predominantly linked to immune responses. The Interferome database confirmed a predominance of type I and II IFN signatures in both groups. Pathway enrichment analysis highlighted inflammatory responses, including cytokine production (IL-8, IL1-β, IFN-γ, β, and α) and toll-like receptor signaling (TLR4, TLR7, TLR8, CD180). Principal component analysis emphasized the IFN signature's discriminative power, particularly in systemic disease-associated uveitis. Machine learning identified IFN-associated genes as robust predictors, while linear discriminant analysis pinpointed CCR2, CD180, GAPT, and PTGS2 as key risk factors in isolated uveitis and CA1, SIAH2, and PGS in systemic disease-associated uveitis. CONCLUSION: These findings highlight IFN-driven imune dysregulation and potential molecular targets for precision therapies in uveitis.
Authors
Munhoz DD, Fonseca DLM, Filgueiras IS, Dias HD, Nakaya HI, et al.
External link
Publication Year
Publication Journal
Associeted Project
Network & Precision Medicine
Lista de serviços
-
StructRNAfinder: an automated pipeline and web server for RNA families prediction.StructRNAfinder: an automated pipeline and web server for RNA families prediction.
-
CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.
-
webCEMiTool: Co-expression Modular Analysis Made Easy.webCEMiTool: Co-expression Modular Analysis Made Easy.
-
Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.
-
Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.
-
OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.
-
Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.
-
Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.
-
Automatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone imagesAutomatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone images
-
Tucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approachTucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approach
-
Ten quick tips for harnessing the power of ChatGPT in computational biologyTen quick tips for harnessing the power of ChatGPT in computational biology