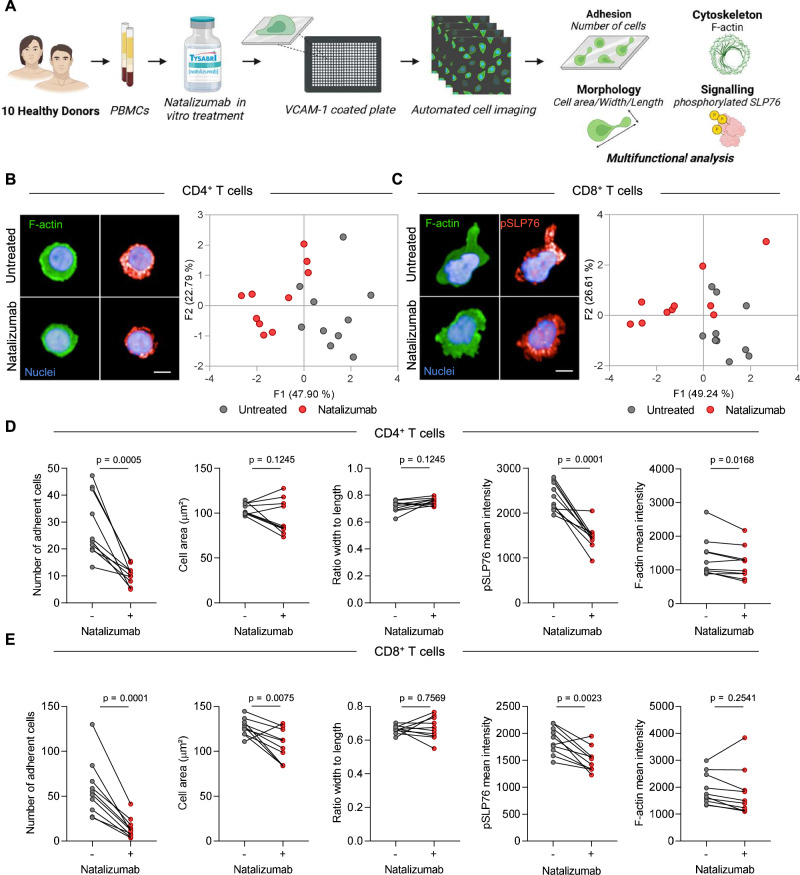
In vitro morphological profiling of T cells predicts clinical response to natalizumab therapy in patients with multiple sclerosis.
Despite the efficacy of natalizumab, which targets the integrin VLA-4, in treating multiple sclerosis (MS), approximately 35% patients with MS presente evidence of disease activity two years after treatment initiation. Individual heterogeneity of leukocyte response to VLA-4 on natalizumab-mediated blockade may underlie disparities in treatment efficacy. Here we use a high-content cell imaging (HCI) pipeline to profile the in vitro effects of natalizumab on VLA-4-stimulated PBMCs from MS patients prior to natalizumab treatment. Unsupervised clustering of image data partially discriminates non-responder MS patients based on morphology, F-actin organization and signaling-related features in CD8+ T cells. Furthermore, through a random forest approach, treatment response can be predicted with a performance of 92% for a Discovery cohort and 88% for a validation cohort. Unfavorable treatment response is associated with a distinct actin remodeling response of natalizumab-exposed CD8+ T cells and a residual ability of these cells to spread on VCAM-1. Our study thus unveils that CD8+ T cells from individual MS patients display heterogeneous susceptibility to natalizumab in vitro and highlights the potential of HCI-based pretreatment monitoring to assist individualized treatment prescription.
Authors
Chaves B, Santos E Silva JC, Nakaya H, Socquet-Juglard N, et al.
External link
Publication Year
Publication Journal
Associeted Project
Network & Precision Medicine
Lista de serviços
-
StructRNAfinder: an automated pipeline and web server for RNA families prediction.StructRNAfinder: an automated pipeline and web server for RNA families prediction.
-
CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.CEMiTool: a Bioconductor package for performing comprehensive modular co-expression analyses.
-
webCEMiTool: Co-expression Modular Analysis Made Easy.webCEMiTool: Co-expression Modular Analysis Made Easy.
-
Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.Assessing the Impact of Sample Heterogeneity on Transcriptome Analysis of Human Diseases Using MDP Webtool.
-
Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.Predicting RNA Families in Nucleotide Sequences Using StructRNAfinder.
-
OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.OUTBREAK: a user-friendly georeferencing online tool for disease surveillance.
-
Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.Noninvasive prenatal paternity determination using microhaplotypes: a pilot study.
-
Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.Editorial: User-Friendly Tools Applied to Genetics or Systems Biology.
-
Automatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone imagesAutomatic detection of the parasite Trypanosoma cruzi in blood smears using a machine learning approach applied to mobile phone images
-
Tucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approachTucuxi-BLAST: Enabling fast and accurate record linkage of large-scale health-related administrative databases through a DNA-encoded approach
-
Ten quick tips for harnessing the power of ChatGPT in computational biologyTen quick tips for harnessing the power of ChatGPT in computational biology