Concepts on Microarray Design for Genome and Transcriptome Analyses
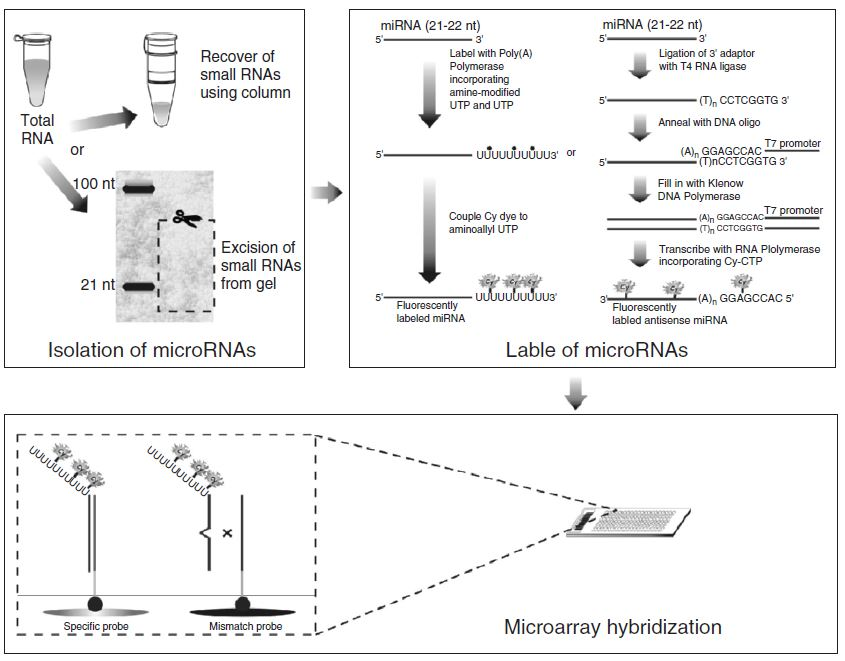
Microarray technology has revolutionized molecular biology by permitting many hybridization experiments to be performed in parallel. With the size of a glass microscope slide, this tool can carry thousands of DNA fragments in an area smaller than a postage stamp. In this chapter, we will describe microarray chips that host nucleic acid probes, which are the most commonly used type of microarrays. Science in this field is mostly data-driven, where biological hypothesis are generated upon analysis and comparison of a huge amount of potentially meaningful differential data derived from microarray hybridizations. DNA microarray technology is under a constant and rapid evolution. The first paper reporting DNA microarray as a tool for transcript-level analyses has been published in 1995, and that chip had about 1000 Arabidopsis genes. Almost 11 years have passed and advances in miniaturization, robotic, and informatics, as well as the development of alternative approaches to microarray construction have permitted to put more than 250,000 different spots into a single square centimeter. This rapid advance in the microarray field, combined with the falling price of technology and the acquisition of wholegenome sequence information for hundreds of organisms has caused biologists to abandon their home-made equipment in favor of one of an expanding range of commercial platforms now available on the market. However, we are still not able to represent the entire genome of any eukaryotic organism in a unique chip or even to analyze the great complexity of the human transcriptome. Therefore, how to choose and design the best probes to construct DNA microarray chips is a crucial step to the appropriate use of this powerful technique (Figure 1).
Authors
Helder I. Nakaya; Eduardo M. Reis; Sergio Verjovski-Almeida
External link
Publication Year
Publication Journal
Associeted Project
Life Sciences
Lista de serviços
-
RASL11A, member of a novel small monomeric GTPase gene family, is down-regulated in prostate tumors.RASL11A, member of a novel small monomeric GTPase gene family, is down-regulated in prostate tumors.
-
Splice variants of TLE family genes and up-regulation of a TLE3 isoform in prostate tumors.Splice variants of TLE family genes and up-regulation of a TLE3 isoform in prostate tumors.
-
Concepts on Microarray Design for Genome and Transcriptome AnalysesConcepts on Microarray Design for Genome and Transcriptome Analyses
-
The iron stimulon of Xylella fastidiosa includes genes for type IV pilus and colicin V-like bacteriocins.The iron stimulon of Xylella fastidiosa includes genes for type IV pilus and colicin V-like bacteriocins.
-
Origins of the Xylella fastidiosa prophage-like regions and their impact in genome differentiation.Origins of the Xylella fastidiosa prophage-like regions and their impact in genome differentiation.
-
The role of prophage in plant-pathogenic bacteria.The role of prophage in plant-pathogenic bacteria.
-
Genetic control of immune response and susceptibility to infectious diseases.Genetic control of immune response and susceptibility to infectious diseases.
-
Building capacity for advances in tuberculosis research; proceedings of the third RePORT international meeting.Building capacity for advances in tuberculosis research; proceedings of the third RePORT international meeting.
-
São Paulo School of Advanced Sciences on Vaccines: an overview.São Paulo School of Advanced Sciences on Vaccines: an overview.
-
A reasonable request for true data sharing.A reasonable request for true data sharing.