Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer.
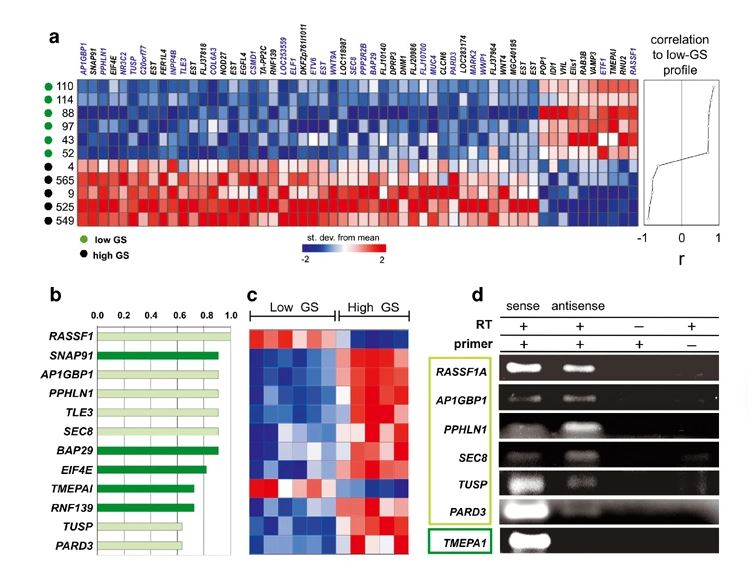
A large fraction of transcripts are expressed antisense to introns of known genes in the human genome. Here we show the construction and use of a cDNA microarray platform enriched in intronic transcripts to assess their biological relevance in pathological conditions. To validate the approach, prostate cancer was used as a model, and 27 patient tumor samples with Gleason scores ranging from 5 to 10 were analyzed. We find that a considerably higher fraction (6.6%, [23/346]) of intronic transcripts are significantly correlated (P< or =0.001) to the degree of prostate tumor differentiation (Gleason score) when compared to transcripts from unannotated genomic regions (1%, [6/539]) or from exons of known genes (2%, [27/1369]). Among the top twelve transcripts most correlated to tumor differentiation, six are antisense intronic messages as shown by orientation-specific RT-PCR or Northern blot analysis with strand-specific riboprobe. Orientation-specific real-time RT-PCR with six tumor samples, confirmed the correlation (P=0.024) between the low/high degrees of tumor differentiation and antisense intronic RASSF1 transcript levels. The need to use intron arrays to reveal the transcriptome profile of antisense intronic RNA in cancer has clearly emerged.
Authors
Eduardo M Reis; Helder I Nakaya; Rodrigo Louro; Flavio C Canavez; Aurea V F Flatschart; Giulliana T Almeida; Camila M Egidio; Apuã C Paquola; Abimael A Machado; Fernanda Festa; Denise Yamamoto; Renato Alvarenga; Camille C da Silva; Glauber C Brito; Sérgio D Simon; Carlos A Moreira-Filho; Katia R Leite; Luiz H Camara-Lopes; Franz S Campos; Etel Gimba; Giselle M Vignal; Hamza El-Dorry; Mari C Sogayar; Marcello A Barcinski; Aline M da Silva; Sergio Verjovski-Almeida
External link
Publication Year
Publication Journal
Associeted Project
Long Noncoding RNAs
Lista de serviços
-
As antisense RNA gets intronic.As antisense RNA gets intronic.
-
Androgen responsive intronic non-coding RNAs.Androgen responsive intronic non-coding RNAs.
-
Conserved tissue expression signatures of intronic noncoding RNAs transcribed from human and mouse loci.Conserved tissue expression signatures of intronic noncoding RNAs transcribed from human and mouse loci.
-
The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation.The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation.
-
Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer.Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer.
-
Insight Into the Long Noncoding RNA and mRNA Coexpression Profile in the Human Blood Transcriptome Upon Leishmania infantum Infection.Insight Into the Long Noncoding RNA and mRNA Coexpression Profile in the Human Blood Transcriptome Upon Leishmania infantum Infection.
-
Long non-coding RNAs associated with infection and vaccine-induced immunityLong non-coding RNAs associated with infection and vaccine-induced immunity
-
Comparative transcriptomic analysis of long noncoding RNAs in Leishmania-infected human macrophagesComparative transcriptomic analysis of long noncoding RNAs in Leishmania-infected human macrophages
-
SARS-CoV-2 Selectively Induces the Expression of Unproductive Splicing Isoforms of Interferon, Class I MHC, and Splicing Machinery Genes.SARS-CoV-2 Selectively Induces the Expression of Unproductive Splicing Isoforms of Interferon, Class I MHC, and Splicing Machinery Genes.