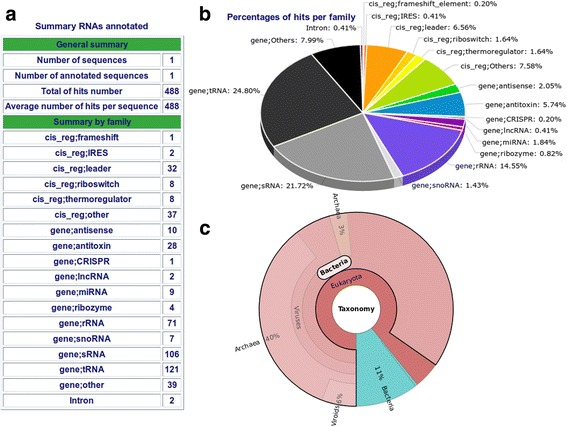
StructRNAfinder: an automated pipeline and web server for RNA families prediction.
The function of many noncoding RNAs (ncRNAs) depend upon their secondary structures. Over the last decades, several methodologies have been developed to predict such structures or to use them to functionally annotate RNAs into RNA families. However, to fully perform this analysis, researchers should utilize multiple tools, which require the constant parsing and processing of several intermediate files. This makes the large-scale prediction and annotation of RNAs a daunting task even to researchers with good computational or bioinformatics skills. We present an automated pipeline named StructRNAfinder that predicts and annotates RNA families in transcript or genome sequences. This single tool not only displays the sequence/structural consensus alignments for each RNA family, according to Rfam database but also provides a taxonomic overview for each assigned functional RNA. Moreover, we implemented a user-friendly web service that allows researchers to upload their own nucleotide sequences in order to perform the whole analysis. Finally, we provided a stand-alone version of StructRNAfinder to be used in large-scale projects. The tool was developed under GNU General Public License (GPLv3) and is freely available at http://structrnafinder.integrativebioinformatics.me . The main advantage of StructRNAfinder relies on the large-scale processing and integrating the data obtained by each tool and database employed along the workflow, of which several files are generated and displayed in user-friendly reports, useful for downstream analyses and data exploration.
Authors
Raúl Arias-Carrasco; Yessenia Vásquez-Morán; Helder I Nakaya; Vinicius Maracaja-Coutinho
External link
Publication Year
Publication Journal
Associeted Project
User-friendly computational Tools
Lista de serviços
-
As antisense RNA gets intronic.As antisense RNA gets intronic.
-
Androgen responsive intronic non-coding RNAs.Androgen responsive intronic non-coding RNAs.
-
Conserved tissue expression signatures of intronic noncoding RNAs transcribed from human and mouse loci.Conserved tissue expression signatures of intronic noncoding RNAs transcribed from human and mouse loci.
-
The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation.The intronic long noncoding RNA ANRASSF1 recruits PRC2 to the RASSF1A promoter, reducing the expression of RASSF1A and increasing cell proliferation.
-
Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer.Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer.
-
Insight Into the Long Noncoding RNA and mRNA Coexpression Profile in the Human Blood Transcriptome Upon Leishmania infantum Infection.Insight Into the Long Noncoding RNA and mRNA Coexpression Profile in the Human Blood Transcriptome Upon Leishmania infantum Infection.
-
Long non-coding RNAs associated with infection and vaccine-induced immunityLong non-coding RNAs associated with infection and vaccine-induced immunity
-
Comparative transcriptomic analysis of long noncoding RNAs in Leishmania-infected human macrophagesComparative transcriptomic analysis of long noncoding RNAs in Leishmania-infected human macrophages
-
SARS-CoV-2 Selectively Induces the Expression of Unproductive Splicing Isoforms of Interferon, Class I MHC, and Splicing Machinery Genes.SARS-CoV-2 Selectively Induces the Expression of Unproductive Splicing Isoforms of Interferon, Class I MHC, and Splicing Machinery Genes.